 |
The big picture
Many species worldwide are experiencing rapid, human-induced changes in their environment that threaten their persistence, but we still struggle to predict which species will adapt to these changing circumstances, and how adaptation may affect species composition in nature. We develop ecological models, evolutionary models, and statistical inference techniques to detect selection signals in genomic and phenotypic data. Long-term goals of our work are to use these models to predict when species maintain sufficient genetic variation to adapt to rapid environmental change, and to understand how evolutionary processes affect our ability to detect the genetic variation underlying heritable traits.
How predictable are eco-evolutionary responses to environmental change?
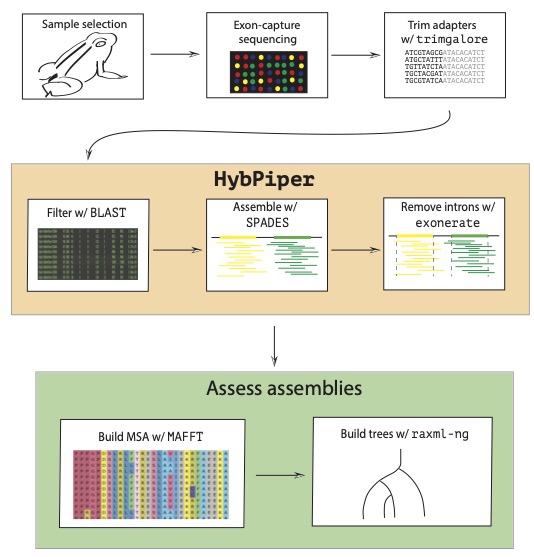
Evolutionary biologists have catalogued many adaptation events and studied their genomic basis across species, such as the convergent evolution of toxicity or rapid
evolution of pesticide resistance.
Most of these inferences of adaptation
proceed backwards in time -- genomic, phenptypic, and ecological data are leveraged to explain how past environments led to the variation that we observe today.
How can we repurpose these backwards-looking methods to understand when adaptation is likely to occur? Is adaptation always inherently unpredictable, or
are there certain traits and environmental changes that result in predictable outcomes in terms of population dynamics or adaptation events?
We see this as both a pressing application of ecological and
evolutionary theory within the context of global change and a fundamental theoretical question about the rate, mode, and mechansism of
adaptive evolution. We are studying questions related to the genomic and evolutionary
response to environmental change
in a number of systems, including marine molluscs, frogs, pigeons, moths, and plants such as alpine carnations.
PhD students SJ McGeady and Alejandro Calderon are both working in this area. SJ is investigating how infectious disease spillover
affects both population dynamics and adaptation rates, specifically in conserved disease-interacting genes.
Alejandro is developing simulation methods to study how changes
in voltinism (the number of generations per year) for insect populations could affect the rate of differentiation in their genomes in collaboration
with Erik Dopman's lab.
Characterizing assembly processes in ecological communities
 Exotic species can dramatically affect species composition by directly out-competing native species, but in some cases native and introduced species may coexist stably.
We studied competition between native and exotic species in California grasslands. Our analysis suggests
that exotic annual species are liekly to outcompete California native species, but that their likelihood of persistence depends integrally on the order of arrival of their exotic competitors.
Our results provide a contrast to some earlier studies that focused on undisturbed communities and found stabilizing niche differences between species, perhaps due to the long co-evolutionary history of naturally co-occurring species. Studies like this one can help inform our thinking about the processes that
could explain diversity in nature, but we still know so little about how biotic interactions affect species' distributions and persistence!
Exotic species can dramatically affect species composition by directly out-competing native species, but in some cases native and introduced species may coexist stably.
We studied competition between native and exotic species in California grasslands. Our analysis suggests
that exotic annual species are liekly to outcompete California native species, but that their likelihood of persistence depends integrally on the order of arrival of their exotic competitors.
Our results provide a contrast to some earlier studies that focused on undisturbed communities and found stabilizing niche differences between species, perhaps due to the long co-evolutionary history of naturally co-occurring species. Studies like this one can help inform our thinking about the processes that
could explain diversity in nature, but we still know so little about how biotic interactions affect species' distributions and persistence!
Our ongoing work seeks to incorporate genomic data into ecological models to predict when species composition is likely to depend on rapid evolutionary
processes. Evolutionary dynamics could be an imporant component of assembly processes in nature, and could be particularly relevant for species that
experience shifts in their environental context due to climate change or introduction to new habitats. Microbes are an ideal model for asking such questions
because of their short generation times, feasibility of whole-genome sequencing, well-characterized biological interactions, and natural hitory relevance.
Graduate student Kasturi Lele is using a combination of experimental microbial community assembly data, genomic sequecnes, and model-based simulations
to characterize how evolutionary and ecological processes may affect community stability in the sourdough microbiome. She is working in collaboration with
Dr. Ben Wolfe's lab at Tufts.