Analyzing the output¶
sfs_code stores a great deal of information about each mutation or substitution in a simulation, which can make it challenging to parse the output. With sfs_coder, all the data is stored internally in the Mutation class, allowing for flexible manipulation of the output.
Opening and reading an output file¶
Opening and reading files is simple. Below is an example that reads in all
the data in an SFS_CODE output file and calculates 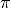 (the average
pairwise diversity) in the 0th locus in all sampled populations.
(the average
pairwise diversity) in the 0th locus in all sampled populations.
from sfscoder import sfs
import sys
# an sfs_code output file that we will analyze
f =sys.argv[1]
# initializing a data object and setting the file path
data = sfs.SFSData(file=f)
# getting all the data from the simulations in the file
data.get_sims()
# The simulations in the file are stored in the data.sims attribute
# for each Simulation object in data.sims, we can calculate pi for
# a set of loci
for sim in data.sims:
pis = sim.calc_pi(loci=[0]) # array of pi values, indexed by population
print pis[0] # pis[0] is pi in the 0th population